BIONUMERICS uses proven technology acquired from the Kodon software to bring various specialized genome comparison and analysis functions. The Genome Analysis Tools module offers side-by-side comparison of genomes and chromosomes, genome annotation, analysis of organization and functional behavior of genomes, alignment of multiple chromosomes and chromosome-based SNP analysis.
This module can only be used in combination with the Sequence Data module.
Features:
Comparative genomics tools
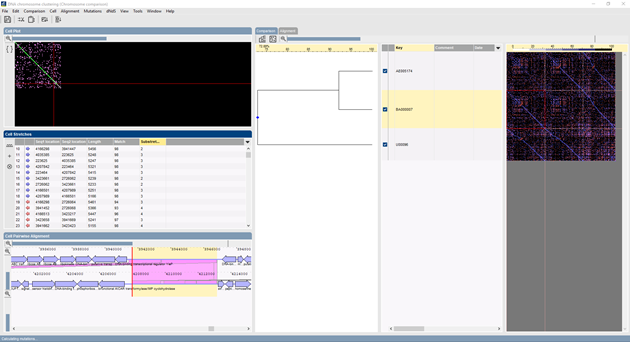
BIONUMERICS lets you align and compare sequences of up to full chromosome length. Discontinuous alignments are calculated using seed and stretch-based sequence mapping, revealing genomic inversions, swaps, duplications, insertions and deletions. Mutation and SNP discovery can be performed on template-based multiple chromosome alignments, with optional selection of mutation type (intergenic, synonymous, non-synonymous or indel) and filtering of significant SNPs based on quality scores. dNdS analysis based on the ratio synonymous/non-synonymous mutations within gene clusters is available to predict evolutionary selection pressure on genes.
Genome annotation
Unannotated or partially annotated sequences can be annotated against one or multiple reference chromosomes. For each annotated ORF, all possible choices are listed according to feature identity and chromosome synteny. Similar features can be clustered according to identity. The user can manually override annotations made by the software.
