Demanding calculations on whole-genome sequencing (WGS) data can be performed on an external Calculation Engine which is completely integrated with the BIONUMERICS client software.
From within the BIONUMERICS software, you can post jobs on the Calculation Engine and easily retrieve the results. This keeps your computer responsive and your database lightweight, so you can focus on the actual analysis. It also means that hardware requirements for your desktop or laptop computer running BIONUMERICS are kept modest and that you can let the Calculation Engine do all the heavy lifting!
What is the Calculation Engine?
The Calculation Engine is a server application that provides easy access from the BIONUMERICS desktop client application to a high-performance computing (HPC) environment for doing calculation-intensive tasks, including de novo assembly, reference mapping and whole genome multi-locus sequence typing (wgMLST) allele calling. A nomenclature service is integrated with the Calculation Engine for wgMLST.
The Calculation Engine is installed on a powerful computer cluster, either locally or in the cloud.
Interaction between BIONUMERICS and the Calculation Engine
All communication between the BIONUMERICS client and the Calculation Engine service is handled by the WGS tools plugin, which is already included in your BIONUMERICS installation. Authentication is done through a project name and password, linked to your BIONUMERICS license.
The WGS tools plugin allows you to post jobs on WGS data to the Calculation Engine and to retrieve the results of these jobs after the calculations are finished. In addition, the plugin
- sets up your database by creating the necessary experiment types,
- retrieves optimal settings for allele calling from the curator database,
- synchronizes with the nomenclature server in wgMLST, and
- provides a mapping of BIONUMERICS wgMLST analyses to public nomenclature (e.g. BIGSdb),
- enables import of sequence read sets as links from the NCBI Sequence Read Archive (SRA), EMBL-EBI European Nucleotide Archive (ENA), Amazon S3 buckets, Illumina BaseSpace or local file servers.
In short, simply installing the WGS tools plugin and entering your credentials to a project unlocks the full power of the Calculation Engine!
The BIONUMERICS Cloud Calculation Engine
The default Calculation Engine instance in the WGS tools plugin is the BIONUMERICS Cloud Calculation Engine, which is hosted on Amazon servers and accessible worldwide. It is designed to process hundreds of isolates within the hour, providing extremely fast turnaround times for the primary analysis in wgMLST and wgSNP.
Leveraging the BIONUMERICS Cloud Calculation Engine for your research requires no investment in computer hardware and is almost setup-free. The only thing you need is a Calculation Engine project, which can be requested online.
Analyses on the BIONUMERICS Cloud Calculation Engine are charged at a pay-per-use system through so-called Calculation Engine credits (1 credit is 1 Euro). Following analyses are offered:
- Reference mapping: 1 credit
- De novo assembly: 1 credit
- De novo assembly (hybrid): 10 credits
- wgMLST assembly-based calls: 3 credits
- wgMLST assembly-free calls: 3 credits
- Annotation by Prokka: 1 credit
- CFSAN SNP pipeline: 5 credits
- RAxML pipeline: 5 credits
- FastTree pipeline: 3 credits
- MTBC genotyping: 2 credits
- SeqSero: 1 credit
Credits for the BIONUMERICS Cloud Calculation Engine can be ordered via our online credits order form.
Data security and the BIONUMERICS Cloud Calculation Engine
We take privacy and data security seriously. Therefore, the communication scheme between BIONUMERICS and the Calculation Engine is based on following principles:
- No metadata is ever sent to the Calculation Engine.
- We always use secure connections (HTTPS) when private data are transmitted.
- All exposed services use strong passwords and encryption algorithms.
For more details, check out our Knowledge Base article on what information is sent to the Calculation Engine.
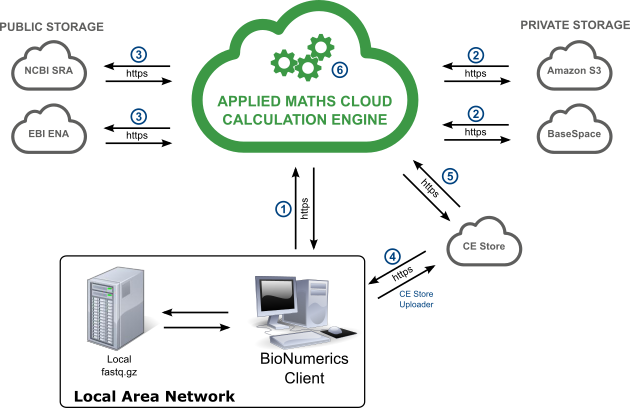
The sequence of actions when running a job on the Calculation Engine is as follows:
- The user launches a job to the Calculation Engine.
- If the sequence reads are stored locally, a separate executable (CEStore Uploader) is started and uploads fastq.gz files to the CE Store (4). No metadata are stored on the CE Store, private storage or public storage!
- Via the sequence read set link, the Calculation Engine fetches the sequence reads from CE Store (5), private storage (2) or public storage (3).
- The HPC cluster on the Calculation Engine executes the job and makes the results available for the BIONUMERICS client.
- The user downloads the results for the submitted job.
The CE Store is Amazon S3 storage managed by the Calculation Engine. This means that only the Calculation Engine can read data from the CE Store and obsolete files are deleted when their caching time is expired.
Local Calculation Engine deployments
With large sample volumes, it can be beneficial to have a local installation of the Calculation Engine, either on your institute’s own computer cluster or as a private cloud solution. Several influential public health institutes and major industrial customers already have their own installation of the Calculation Engine.
For local Calculation Engine deployments, we provide installation either on-site or remotely. The calculation engine installation, maintenance and updates are defined in customized service contracts.
Local Calculation Engine deployments can also provide an outcome for institutes facing strict local or national data protection laws, excluding the use of any public or private cloud services. With a Calculation Engine installed on a local computer cluster, your data effectively does not leave the premises!
Contact us for more information and a price quotation.
